iscream currently has Rhtslib as a “LinkingTo” dependency so if a
system installation of htslib is not found with pkg-config, the
installer will fall back to using Rhtslib as the htslib header source.
However, we recommend getting a more up-to-date htslib from another
source. You or your system administrator can install htslib with the
system package manager which usually sets PKG_CONFIG_PATH
automatically. On MacOS you can get htslib with the Homebrew package
manager. On HPC systems, htslib may be provided as a module. Make sure
these methods also set the PKG_CONFIG_PATH.
If you aren’t able to install htslib development libraries system-wide for lack of admin permissions, you can install them from other channels. We recommend pixi or conda since their htslib is compiled with libdeflate support and is faster than htslib without libdeflate. If you’re compiling your own htslib, compile libdeflate first and then htslib. With Rhtslib we’ve seen poorer performance compared to a standard htslib installation, both with and without libdeflate.
To see what htslib version iscream is using and whether it has libdeflate, run
library(iscream)
htslib_version()
#> 1.21
#> build=configure libcurl=yes S3=yes GCS=yes libdeflate=yes lzma=yes bzip2=yes plugins=no htscodecs=1.6.1and check that libdeflate=yes.
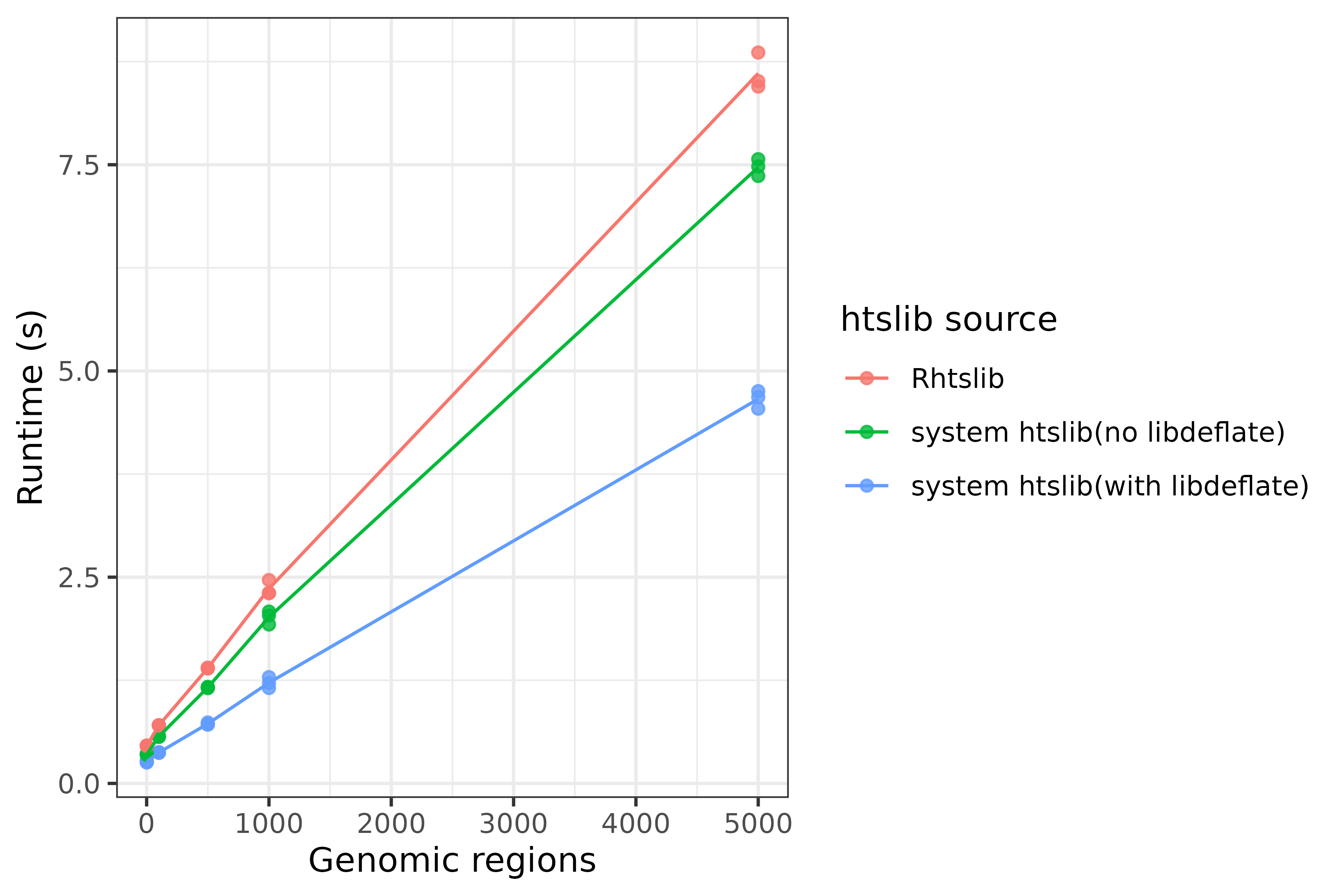
Effect of htslib v1.18 source on iscream’s ‘make_mat()’ runtime from one bulk WGBS BED file
Conda/miniconda/mamba/micromamba
Create an environment.yaml with the following contents
to install htslib 1.21:
Add this file to your project directory and run
conda env create -f environment.yaml
conda activate iscream
export PKG_CONFIG_PATH=$CONDA_PREFIX/lib
export LD_LIBRARY_PATH=$CONDA_PREFIX/lib/Confirm that the headers are available for compilation
You should get something like
-I/home/user/miniconda3/envs/iscream/include -L/home/user/miniconda3/envs/iscream/lib -lhtsPixi
Pixi uses the conda repositories to install packages. Create a
pixi.toml file with this content and add the file to your
project directory:
[project]
channels = ["conda-forge", "bioconda"]
name = "iscream"
platforms = ["linux-64"]
version = "0.1.0"
[activation.env]
LD_LIBRARY_PATH="$CONDA_PREFIX/lib"
[dependencies]
htslib = "1.21.*"
pkg-config = ">=0.29.2,<0.30"To create an environment with the required system dependencies run
Confirm that the headers are available for compilation
You should get something like
-I/home/user/iscream/.pixi/envs/default/include -L/home/user/iscream/.pixi/envs/default/lib -lhtsSession info
## R version 4.5.1 (2025-06-13)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_4.0.0 BiocStyle_2.36.0
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.6 jsonlite_2.0.0 dplyr_1.1.4
## [4] compiler_4.5.1 BiocManager_1.30.26 tidyselect_1.2.1
## [7] jquerylib_0.1.4 systemfonts_1.3.1 scales_1.4.0
## [10] textshaping_1.0.4 yaml_2.3.10 fastmap_1.2.0
## [13] R6_2.6.1 labeling_0.4.3 generics_0.1.4
## [16] knitr_1.50 tibble_3.3.0 bookdown_0.45
## [19] desc_1.4.3 bslib_0.9.0 pillar_1.11.1
## [22] RColorBrewer_1.1-3 rlang_1.1.6 cachem_1.1.0
## [25] xfun_0.53 fs_1.6.6 sass_0.4.10
## [28] S7_0.2.0 cli_3.6.5 pkgdown_2.1.3
## [31] withr_3.0.2 magrittr_2.0.4 digest_0.6.37
## [34] grid_4.5.1 lifecycle_1.0.4 vctrs_0.6.5
## [37] evaluate_1.0.5 glue_1.8.0 farver_2.1.2
## [40] ragg_1.5.0 rmarkdown_2.30 tools_4.5.1
## [43] pkgconfig_2.0.3 htmltools_0.5.8.1